Alternative Splicing
What is alternative splicing?
The human genome contains 19,000 to 20,000 protein coding genes. But here is what is intriguing: There are about 80,000 to 100,000 distinct proteins encoded by this same amount of gene.
How so?
Alternative splicing!
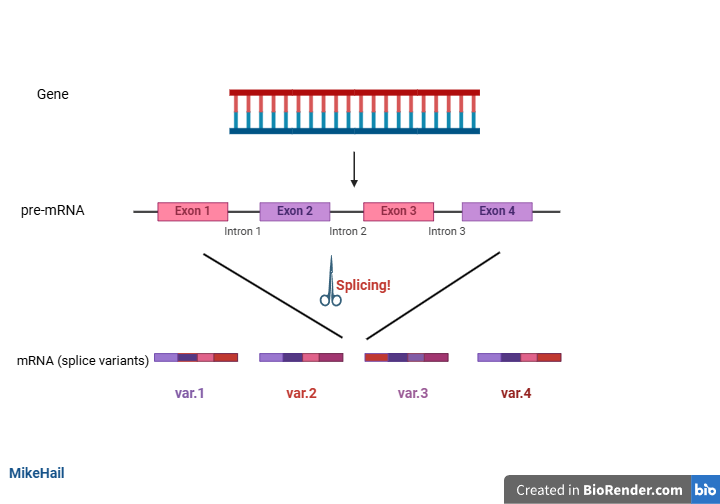
Alternative splicing is simply the mechanism by which a single gene produces multiple mRNAs (splice variants).
Most students of biology agrees with the central dogma of biology: DNA is transcribed to RNA and RNA is translated to protein. The process is more complex. However, the dogma is a good way to start off explaining what alternative splicing means.
As said earlier, there are about 19,000 to 20,000 protein-coding genes. On the other hand, there are approximately 80,000 to 100,000 distinct proteins in humans. From these numbers, it is glaring that the gene-protein relationship is not always 1:1. A gene can produce more than one protein. Alternative splicing involves the splicing (cutting out) of introns from the pre-mRNA. Introns are non-coding regions of pre-mRNAs. They are embedded in between exons, regions which codes for proteins. The splicing process is mediated by molecular machines called the spliceosome. This molecular machinery performs recombination on exons to generate diverse mRNA from a single pre-mRNA, and these molecules are found within the nucleus. The recombination mediated by this molecular machinery are dependent on cell type, developmental changes, or external cues. The introns are spliced out, while exons of choice are selected and rejoined to give multiple mRNA variants, which are then translated into variant proteins. These proteins are from a single gene. However, they are functionally distinct.
Biological importance
- Adaptation to change in external conditions.
In a study done by some researchers on cold acclimation in certain fish (Atlantic killifish, threespine stickleback, and zebrafish), it was observed that cold acclimation led to a substantial change in splicing patterns. The alternative spliced genes were discovered to have been involved in cellular processes previously involved in regulating phenotypic plasticity in response to cold climates (Verhage et al., 2017).
- Disease mechanisms
In many human diseases, defects in splicing have been implicated. For example, in neurofibromatosis, a neurodegenerative disease, an experiment done on mice shows that a defect in alternative splicing which causes increased inclusion of exon 23a contributed to deficits in learning and memory (Mader & Lou, 2022).
Conclusion
Alternative splicing is an important molecular machinery. It allows for diversity in protein production from a single gene. Studying alternative splicing gives an avenue to explore physiological mechanisms. It aids our understanding of what is going wrong in diseased conditions and help scientists come up with therapeutic designs to combat diseases.
References
Verhage, L., Severing, E. I., Bucher, J., Lammers, M., Busscher-Lange, J., Bonnema, G., Rodenburg, N., Proveniers, M. C. G., Angenent, G. C., & Immink, R. G. H. (2017). Splicing-related genes are alternatively spliced upon changes in ambient temperatures in plants. PLoS ONE, 12(3), e0172950. https://doi.org/10.1371/journal.pone.0172950
Mader, K. A., & Lou, H. (2022). Alternative Splicing of Neurofibromatosis Type 1 Exon 23a Modulates Ras/ERK Signaling and Learning Behaviors in Mice. In IntechOpen eBooks. https://doi.org/10.5772/intechopen.99678